Agricultural and Biological Research
RNI # 24/103/2012-R1
Research Article - (2024) Volume 40, Issue 5
Recently, there has been a significant accumulation of data focused on predicting and preventing soybean infections. The methodologies and development stages of these approaches differ, but they share a common goal: To improve crop and product management. This study evaluates various models for disease detection and pod counting in soybean plants. Two specific models, CNN and EfficientDet B0, have been developed to identify healthy and diseased leaves and accurately count pods. Tensor flow, a versatile tool for numerical computation, was utilized in this research. It proves particularly useful in controlled farm environments, where it can quickly detect early signs of disease on plant leaves.
Artificial intelligence; Convolutional Neural Network (CNN); Deep learning; Crop disease; Leaf; Soybean pod
Agriculture is a key source of livelihood. Agriculture provides employment opportunities for village people on a large scale in developing countries like India. India's agriculture is composed of many crops and according to a survey nearly 70% of the population depends on agriculture [1]. Most Indian farmers are adopting manual cultivation due to lagging technical knowledge. Farmers are unaware of what kind of crops that grows well on their land. Plants suffering from different leaf diseases can significantly impact agriculture production, leading to reduced quality and quantity of crops and financial loss. Healthy leaves are crucial for the rapid growth of plants and the enhancement of crop yield. It is difficult for farmers and researchers to identify diseases affecting plant leaves. Presently, farmers resort to spraying pesticides on plants, which can have direct or indirect negative effects on human health and the economy. Rapid detection techniques for plant diseases need to be implemented to address this issue.
Thus detection of plant diseases plays a significant key role within the arena of agriculture. Indian agriculture consists of many crops like cotton, wheat, and soybean. Indian farmers conjointly grow sugarcane, oilseeds, potatoes and non-food things like occasional tea, cotton, rubber. Of these crops support the strength of leaves and roots (Figure 1) [2].

Figure 1 : Infected leaf
Different factors contribute to specific diseases in plant leaves, causing harm to crops and ultimately affecting the economy of the country. Detecting plant diseases early is essential to minimize major losses. Farmers encounter difficulties in recognizing these diseases, frequently lacking the required knowledge for preventive actions. The medical field is crucial in identifying and controlling plant diseases, providing necessary solutions for sustainable agriculture and economic security (Figure 2).
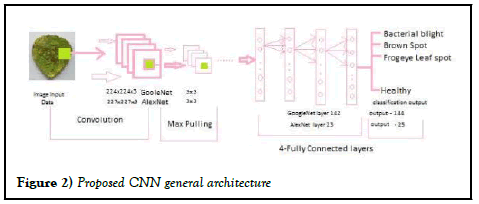
Figure 2: Proposed CNN general architecture
In today's context, image processing methods have emerged as a suitable, cost-effective, and dependable approach for disease detection using plant leaf images (Figure 3).
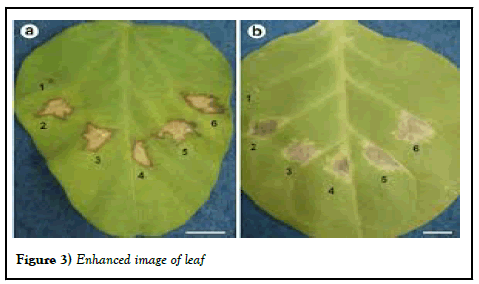
Figure 3: Enhanced image of leaf
Farmers look for economical and effective solutions. Seventy percent of people in the mostly agrarian country of India work in agriculture. Maintaining the caliber and volume of agricultural outputs depends on choosing the right pesticides and treating plant illnesses. Examining discernible patterns on plants is strongly related to the research of plant diseases. Proper monitoring and diagnosis of plant health and illnesses are essential for agricultural production to be effective. In the past, this work required a lot of labor and manual examination by knowledgeable field workers. Such a strategy required a large investment of time and money. Since agriculture continues to be the foundation of the Indian economy, it is critical to encourage sustainable agricultural practices by developing more streamlined and effective disease management techniques.
Using the right methods to distinguish between healthy and sick leaves can help reduce crop loss and boost output. This section includes the many machine-learning methods now in use for diagnosing plant diseases.
Shape and texture-based identification
In Mokhtar U, et al., the authors recognized diseases utilizing tomato-leaf pictures [3,4]. They utilized distinctive mathematical and histogram-based provisions from segmented infected portions and applied a SVM classifier with various portions for characterization. Kaur S, et al., recognized three diverse soybean infections utilizing distinctive shading and surface provisions [5]. In Babu MP, et al., utilized a feed-forward neural organization and back propagation to distinguish plant leaves and their diseases [6]. Chouhan SS, et al., utilized a bacterial-scrounging streamlining based spiral basis function neural organization (BRBFNN) for the distinguishing proof of leaves and contagious diseases in plants [7]. In their methodologies, they utilized a district developing calculation to separate elements from a leaf based on seed focuses having similar attributes. The bacterial-scrounging streamlining procedure is utilized to accelerate organize and further develop arrangement exactness.
Deep-learning-based identification
Mohanty et al., utilized AlexNet and GoogleNet CNN structures in the distinguishing proof of 26 distinctive plant diseases [8]. Ferentinos et al., utilized distinctive CNN structures to recognize 58 diverse plant infections, accomplishing significant degrees of arrangement precision [9]. In their methodology, they additionally tried the CNN design with ongoing pictures. Sladojevic et al., planned a DL design to recognize 13 distinctive plant infections [10]. They utilized the Caffe DL system to perform CNN preparing. Kamilaris et al., comprehensively investigated distinctive DL draws near and their downsides in the field of agribusiness [11]. In Geetharamani G, et al., the creators proposed a nine-layer CNN model to distinguish plant diseases [12]. For experimentation purposes, they utilized the plant village dataset and information expansion methods to expand the information size and investigated execution. The author’s detailed preferable exactness over that of a customary AI based methodology.
Pre-trained AlexNet and GoogleNet were utilized by Jadhav, S.B et al., to identify 3 distinct soybean diseases from solid leaf pictures with adjusted hyper parameters, for example, minibatch size, max age, and inclination learning rate [13]. Six unique pre-prepared network (AlexNet, VGG16, VGG19, GoogleNet, ResNet101 and DenseNet201) utilized by KR Aravind et al., to distinguish 10 unique infections in plants, and they accomplished the most elevated exactness pace of 97.3% utilizing GoogleNet [14]. A pretrained VGG16 as the component extractor and multiclass SVM were utilized by Rangarajan AK, et al., to arrange diverse eggplant disease [15]. Distinctive shading spaces (RGB, HSV, YCbCr, and grayscale) were utilized to assess execution; utilizing RGB pictures, the most noteworthy characterization precision of 99.4% was accomplished. In Arora J, et al., the creators grouped maize leaf disease from solid leaves utilizing profound woodland strategies [16]. In their methodology, they fluctuated the deep woodland hyper parameters in regards to number of trees, timberlands, and grains, what's more, contrasted their outcomes and those of customary AI models, for example, SVM, RF, LR, and KNN. Lee et al., looked at changed profound learning structures in the ID of plant infections [17]. To work on the precision of the model, Ghazi et al., utilized an exchange learning-put together methodology with respect to pre-trained deep learning models [18].
In Li Y, et al., utilized a shallow CNN with SVM and RF classifiers to group three unique kinds of plant infections [19]. In their work, they primarily analyzed their outcomes with those of deep learning strategies and showed that arrangement utilizing SVM and RF classifiers with removed components from the shallow CNN beat pre-trained deep learning models. A self-consideration convolutional neural organization (SACNN) was utilized by Zeng W, et al., to recognize a few yield diseases [20]. To inspect the power of the model, the creators added diverse commotion levels in the test-picture set. Oyewola et al., recognized 5 diverse cassava-plant diseases utilizing plain convolutional neural organization (PCNN) and Deep Residual Neural Network (DRNN), and found that DRNN outflanked PCNN by an edge of 9.25% [21]. Ramacharan et al., utilized an exchange learning approach in the distinguishing proof of three infections and two nuisance harm types in cassava plants. The creators then, at that point, broadened their work on the ID of cassava plant infections utilizing a cell phone based CNN model and accomplished precision of 80.6%.
A NASNet-based profound CNN design was utilized by Mohanty et al., to distinguish leaf infections in plants, and a precision pace of 93.82% was accomplished. Rice-and maize-leaf infections were recognized by Chen et al., utilizing the INC-VGGN strategy [22]. In their methodology, they supplanted the last convolutional layer of VGG19 with two initiation layers and one worldwide normal pooling layer. A shallow CNN (SCNN) was utilized by Yang Li et al., in the ID of maize, apple, and grape illnesses. In the first place, they separated CNN highlights and ordered them utilizing SVM and RF classifiers. Sethy et al., utilized distinctive deep learning models to remove includes and arrange them utilizing a SVM classifier [23]. Utilizing ResNet50 with SVM, they accomplished the best precision. A VGG16, ResNet, and DenseNet model was utilized by Yafeng Zhao et al., to recognize plant illnesses from the plant village dataset [24,25]. To expand the dataset size, they utilized a generative adversarial network (DoubleGAN), which further developed the presentation results. A summary of the related work on plant-diseases dependent on leaf.
Convolutional Neural Networks (CNNs) have advanced significantly in image categorization and object recognition. In the past, learning algorithms were applied to feature spaces containing hand-crafted features such as SURF, HoG, and SIFT, among others, in order to classify images. These techniques relied on well stated predetermined features to work. Nevertheless, feature engineering is a laborious and intricate procedure that must be repeated each time the dataset or problem undergoes a substantial alteration. This constraint is apparent in conventional computer visionbased plant disease detection efforts, which mostly rely on labor-intensive processes such as picture augmentation and hand-crafted features. Using infected plant leaves, a variety of techniques have been developed for the identification and categorization of plant diseases. Unfortunately, there haven't been many effective and profitable commercial disease identification methods. In order to identify plant illnesses in soybean leaves and pods, we used a variety of Deep Learning (DL) models in our study, such as convolutional neural networks and EfficientNetB0. We obtained data from ICAR (Indian Institute-Soybean Research) for the purpose of training and testing the model. This dataset comprised a standard set of 200 photos showing soybean pods plus 1951 images showing healthy and sick soybean leaves from 14 different species. The training and testing portions of the dataset were split 60-40, and the EfficientNetB0 model produced an astounding accuracy rate of 99.56%. Additionally, when working with colored images, our training procedure was effective, requiring 565 and 545 seconds per epoch for CNN and EfficientNetB0, respectively. Our applied model performs better in terms of prediction than other deep learning techniques, providing higher accuracy and lower loss. Plant disease identification is now much more accurate and efficient thanks to developments in deep learning models, which also present a promising future for agricultural applications. Plant disease diagnosis could be revolutionized with the application of CNNs and models such as EfficientNetB0. This could result in more timely and effective interventions, which could ultimately improve crop output and food production.
Mr. Nitesh Rastogi is a Research Scholar, Faculty of Engineering and Technology, for his renowned and sound research in this particular area is a spotlight in the field of agriculture.
Dr. Priya R. Swaminarayan, Dean, Faculty of IT and Computer Science, for her benevolent and constant support as a mentor for the fulfillment of this research which is a milestone the field of agriculture in combination with IT and computer science.
ORCID ID: https://orcid.org/0000-0001-5113-1922
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Rastogi N, Swaminarayan PR. EfficientNetB0 technique of CNN for disease and health identification of soybean plant. AGBIR.2024;40(5): 1-3.
Received: 02-Sep-2024, Manuscript No. AGBIR-24-147134; , Pre QC No. AGBIR-24-147134 (PQ); Editor assigned: 05-Sep-2024, Pre QC No. AGBIR-24-147134 (PQ); Reviewed: 19-Sep-2024, QC No. AGBIR-24-147134; Revised: 09-Oct-2024, Manuscript No. AGBIR-24-147134 (R); Published: 16-Oct-2024, DOI: 10.37532/0970-1907.24.40(5):1335-1337
